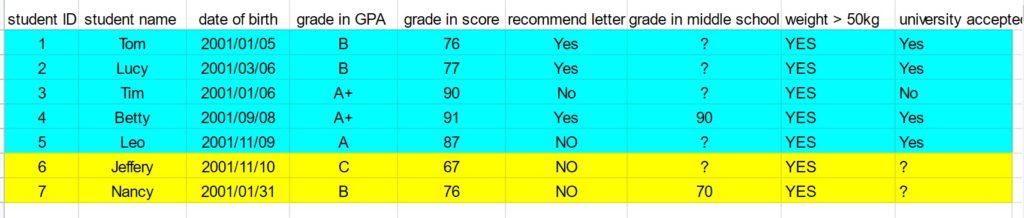
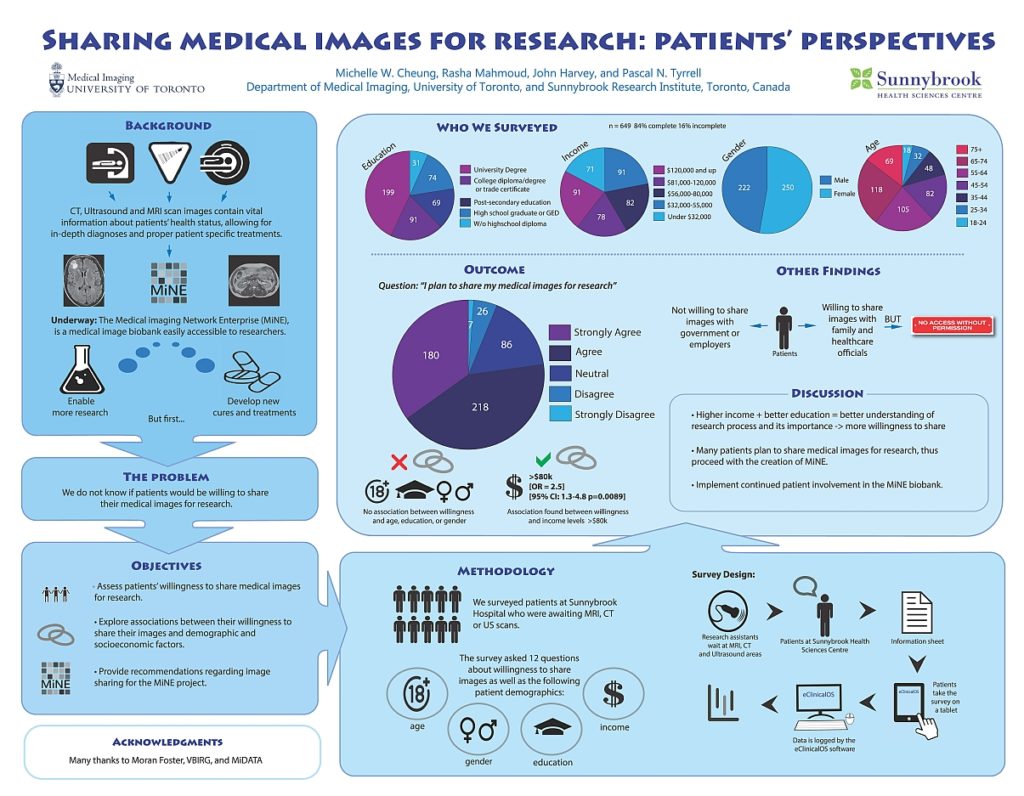
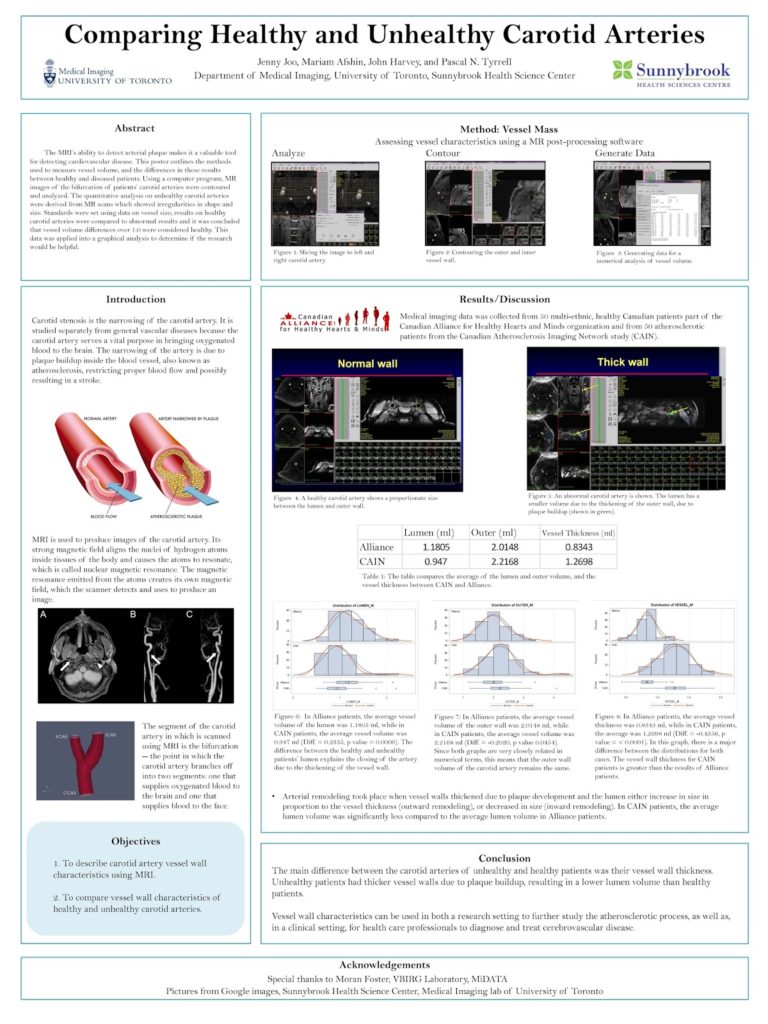
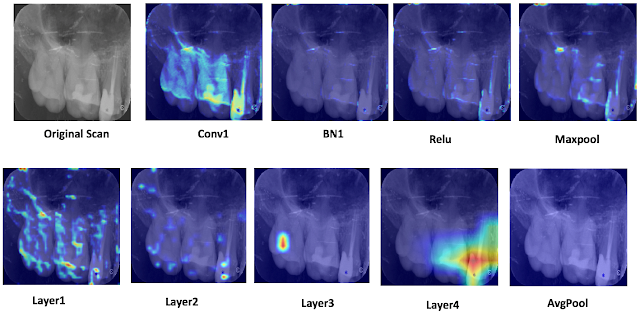
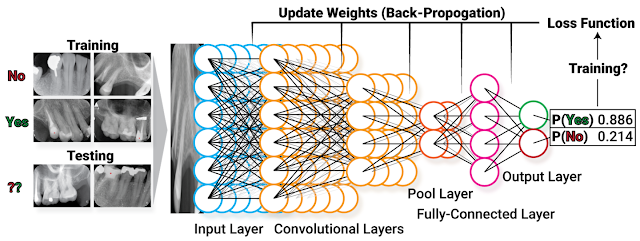
My name is Jacky Wang, and I am just finishing my third year at the University of Toronto, pursuing a computer science specialist. Looking back on this challenging but incredible year, I was honoured to have the opportunity to work inside Dr. Tyrrell’s lab as part of the ROP399 course. I would love to share my experience studying and working inside the lab.
Looking back, I realize one of the most challenging tasks is getting onboard. I felt a little lost at first when surrounded by loads of new information and technologies that I had little experience with before. Though feeling excited by all the collision of ideas during each meeting, having too many choices sometimes could be overwhelming. Luckily after doing more literature review and with the help of the brilliant researchers in the lab (a big thank you to Mauro, Dimitri, and of course, Dr. Tyrrell), I start to get a better view of the trajectories of each potential project and further determine what to get out from this experience. I did not choose the machine learning projects, though they were looking shiny and promising as always (as a matter of fact, they turned out to be successful indeed). Instead, I was more leaning towards studying the sample size determination methodology, especially the concept of ill-posed problems, which often occur when the researchers make conclusions from models trained on limited samples. It had always been a mystery why I would get different and even contrasting results when replicating someone else’s work on smaller sample sizes. From there, I settled the research topic and moved onto the implementation details.
This year the ROP students are coming from statistics, computer science and biology etc. I am grateful that Dr. Tyrrell is willing to give anyone who has the determination to study in his lab a chance though they may have little research experience and come from various backgrounds. As someone who studies computer science with a limited statistics background, the real challenge lies in understanding all the statistical concepts and designing the experiments. We decided to apply various dimension reduction techniques to study the effect of different sample sizes with many features. I designed experiments around the principal component analysis (PCA) technique while another ROP student Jessica explored the lasso and SES model in the meantime. It was for sure a long and memorable experience with many debugging when implementing the code from scratch. But it was never more rewarding than seeing the successful completion of the code and the promising results.
I feel lucky and grateful that Dr. Tyrell helped me complete my first research project. He broke down the long and challenging research task into clear and achievable subgoals within our reach. After completing each subgoal, I could not even believe it sent us close to the finished line. It felt so different taking an ROP course than attending the regular lessons. For most university courses, most topics are already determined, and the materials are almost spoon-fed to you. But sometimes, I start to lose the excitement of learning new topics, as I am not driven by the curiosity nor the application needs but the pressure of being tested. However, taking the ROP course gives me almost complete control of my study. For ROP, I was the one who decides what topics to explore, how to design the experiment. I could immediately test my understanding and put everything I learned into real applications.
I am so proud of all the skills that I have picked up in the online lab during this unique but special ROP experience. I would like to thank Dr. Tyrrell for giving me this incredible study experience in his lab. There are so many resources out there to reach and so many excellent researchers to seek help from. I would also like to thank all members of the lab for patiently walking me through each challenge with their brilliant insights.
Jacky Wang