Hey! I am Mason Hu, a Data Science Specialist and Math Applications in Stats/Probabilities Specialist who just finished my second year. This summer’s ROP journey in MiDATA lab has been an enlightening journey for me, marking my first formal venture into the world of research. Beyond gaining insight into the intricate technicalities of machine learning and medical imaging, I’ve gleaned foundational lessons that shaped my understanding of the research process itself. My experience can be encapsulated in the following three points:
Research is a journey that begins with a wide scope and gradually narrows down to a focused point. When I was writing my project proposal, I had tons of ideas and planned to test multiple hypotheses in a row. Specifically, I envisioned myself investigating four different attention mechanisms of UNet and assessing all the possible combinations of them, which was already discouraged by Prof. Tyrrell in the first meeting. My aspirations proved to be overambitious, as the dynamic nature of research led me to focus on some unexpected yet incredible discoveries. One example of this would be my paradoxical discovery that attention maps in UNets with residual blocks have almost completely opposite weights to those without. Hence, for a long time, I delved into the gradient flows in residual blocks and tried to explain the phenomenon. Even when time is limited and not all ambitious goals can be reached, the pursuit of just one particular aspect can lead to spectacular insights.
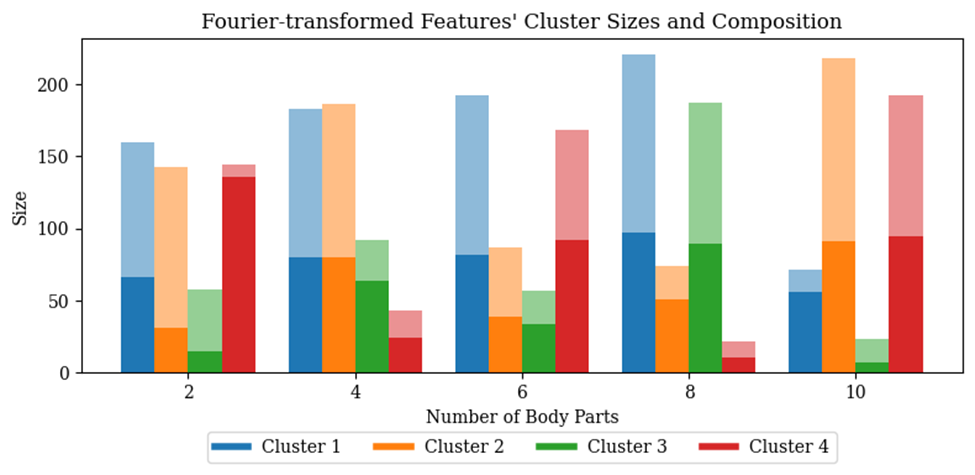
Sometimes plotting out the weights and visualizing them gives me the best sparks and intuitions. This is not restricted to visualizing attention maps in this case. The practice of printing out important statistics and milestones in training models might usually yield great fruition. I once printed out each and every one of the segmentation IoUs in a validation data loader, and it surprised me that some of them are really close to zero. I tried to explain this anomaly as model inefficacy, but it just made no sense. Through an intensive debugging session, I came to realize that it is actually a PyTorch bug specific to batch normalization when the batch size is one. As I go deeper and deeper into the research, I get a better and better understanding of the technical aspects of machine learning and discover better what my research objectives and my purpose are.
Making models reproducible is a really hard task, especially when configurations are complicated. In training a machine learning model, especially CNNs, we usually have a dozen tunable hyperparameters, sometimes more. The technicality of keeping track of them and changing them is already annoying, let alone reproducing them. Moreover, changing an implementation to an equivalent form might not always produce completely equivalent results. Two seemingly equivalent implementations of a function might have different implicit triggers of functionalities that are hooked to one but not the other. This can be especially pronounced in optimized libraries like PyTorch, where subtle differences in implementation can lead to significantly divergent outcomes. The complexity of research underscores the importance of meticulous tracking and understanding of every aspect of the model, affirming that reproducibility is a nuanced and demanding facet of machine learning research.
Reflecting on this summer’s research, I am struck by the depth and breadth of the learning that unfolded. I faced a delicate balance between pursuing big ideas and focusing on careful investigation, always keeping an eye on the small details that could lead to surprising insights. Most importantly, thanks to Prof. Tyrrell, Atsuhiro, Mauro, and Rosa for all the feedback and guidance. Together, they formed a comprehensive research experience for me. As I look to the future, I know that these lessons will continue to shape my thinking, guiding my ongoing work and keeping my curiosity alive.