Ok, a what Transform now??
In the early 1800s, Jean-Baptiste Joseph Fourier, a French mathematician and physicist, introduced the transform in his study of heat transfer. The idea seemed preposterous to many mathematicians at the time, but it has now become an important cornerstone in mathematics.
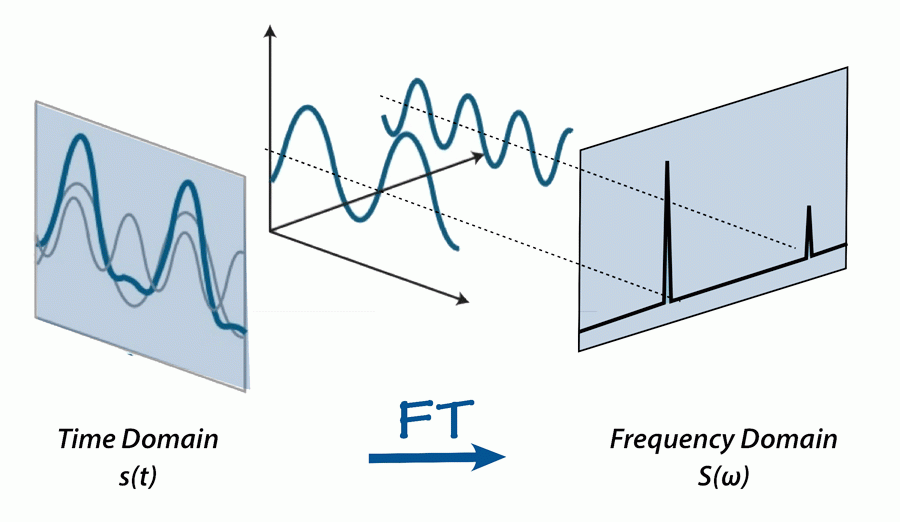
So, what exactly is the Fourier Transform? The Fourier Transform is a mathematical transform that decomposes a function into its sine and cosine components. It decomposes a function depending on space or time into a function depending on spatial or temporal frequency.
Before diving into the mathematical intricacies of the Fourier Transform, it is important to understand the intuition and the key idea behind it. The main idea of the Fourier Transform can be explained simply using the metaphor of creating a milkshake.
Imagine you have a milkshake. It is hard to look at a milkshake and understand it directly; answering questions such as “What gives this shake its nutty flavour?” or “What is the sugar content of this shake?” are harder to answer when we are simply given the milkshake. Instead, it is easier to answer these questions by understanding the recipe and the individual ingredients that make up the shake. So, how exactly does the Fourier Transform fit in here? Given a milkshake, the Fourier Transform allows us to find its recipe to determine how it was created; it is able to present the individual ingredients and the proportions at which they were combined to make the shake. This brings up the questions of how does the Fourier transform determine the milkshake “recipe” and why would we even use this transform to get the “recipe”? To answer the former question, we are able to determine the recipe of the milkshake by running it through filters that then extract each individual ingredient that makes up the shake. The reason we use the Fourier Transform to get the “recipe” is that recipes of milkshakes are much easier to analyze, compare, and modify than working with the actual milkshake itself. We can create new milkshakes by analyzing and modifying the recipe of an existing milkshake. Finally, after deconstructing the milkshake into its recipe and ingredients and analyzing them, we can simply blend the ingredients back to get the milkshake.
Extending this metaphor to signals, the Fourier Transform essentially takes a signal and finds the recipe that made it. It provides a specific viewpoint: “What if any signal could be represented as the sum of simple sine waves?”.
By providing a method to decompose a function into its sine and cosine components, we can analyze the function more easily and create modifications as needed for the task at hand.
A common application of the Fourier Transform is in sound editing. If sound waves can be separated into their “ingredients” (i.e., the base and treble frequencies), we can modify this sound depending on our requirements. We can boost the frequencies we care about while hiding the frequencies that cause disturbances in the original sound. Similarly, there are many other applications of the Fourier Transform such as image compression, communication, and image restoration.
This is incredible! An idea that the mathematics community was skeptical of, now has applications to a variety of real-world applications.
Now, for the fun part, using Fourier Transform in a sentence by the end of the day:
Example 1:
Koby: “This 1000 puzzle is insanely difficult. How are we ever going to end up with the final puzzle picture?”
Eng: “Don’t worry! We can think of the puzzle pieces as being created by taking the ‘Fourier transform’ of the puzzle picture. All we have to do now is take the ‘inverse Fourier Transform’ and then we should be done!”
Koby: “Now when you put it that way…. Let’s do it!”
Example 2:
Grace: “Hey Rohan! What’s the difference between a first-year and fourth-year computer science student?
Rohan: “… what?”
Grace: “A Fouri-y-e-a-r Transform”
Rohan: “…. (╯°□°)╯︵ ┻━┻ ”
I’ll see you in the blogosphere…
Parinita Edke