Having just finished my intro to psychology course, I found that a useful way to learn the material (and guarantee a higher score on the test) was giving myself pop quizzes. In particular, I would write out a phrase and erase a keyword, then I will try to fill in the blank. Turns out this is such an amazing study method that even machines are using it! This method of learning, where the learner learns more in-depth by creating a quiz (e.g. fill-in-the-blank) for itself constitutes the essence of self-supervised learning.
Many machine learning models follow the encoder-decoder architecture, where an encoder network first extracts useful representations from the input data and the decoder network then uses the extracted representations to perform some task (e.g. image classification, semantic segmentation). In a typical supervised learning setting, when there is a large amount of data but only little of which is labelled, all the unlabelled data would have to be discarded as supervised learning requires a model be trained using input-label pairs. On the other hand, self-supervised learning utilizes the unlabelled data by introducing a pretext task to first pretrain the encoder such that it extracts richer and more useful representations. The weights of the pretrained encoder can then be transferred to another model, where it is fine-tuned using the labelled data to perform a specified downstream task (e.g. image classification, semantic segmentation). The general idea of this process is that better representations can be learned through mass unlabelled data, which provides the decoder with a better starting point and ultimately improves the model’s performance on the downstream task.
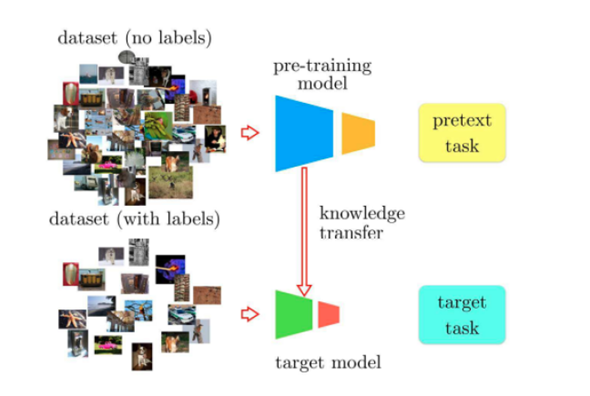
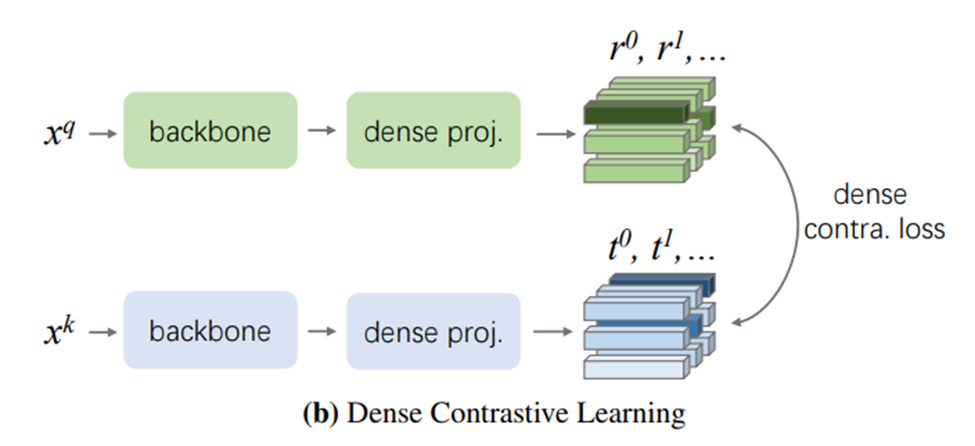
The choice of pretext task is paramount for pretraining the encoder as it decides what kind of representations can be extracted. One powerful pretrain method that has yielded higher downstream performance on image classification tasks is SimCLR. In the SimCLR framework, a batch of images are first sampled, and each image is applied two different augmentations and , resulting in images. Two augmented versions of the same image is called a positive pair, and two augmented versions of different images is called a negative pair. Each pair of the images is passed to the encoder to produce representations and , and these are then passed to a projection layer to obtain the final representations and . A contrastive loss defined using cosine similarity operates on the final representations such that the encoder would produce similar representations for positive pairs and dissimilar representations for negative pairs. After pretraining, the weights of the encoder could then be transferred to a downstream image classification model.

Although better representations may be extracted by the encoder using self-supervised learning, it does require a large unlabelled dataset (typically >100k images for vision-related tasks).
Now, using self-supervised learning in a sentence:
Serious: Self-supervised learning methods such as SimCLR has shown to improve downstream image classification task performance.
Less Serious: I thought you will be implementing a self-supervised learning pipeline for this project, why are you teaching the machine how to solve a rubik’s cube instead? (see Self-supervised Feature Learning for 3D Medical Images by Playing a Rubik’s Cube)
See you in the blogosphere!
Paul Tang