My name is Stanley Hua, and I’ve just finished my 2nd year in the bioinformatics program. I have also just wrapped up my ROP299 with Professor Pascal. Though I have yet to see his face outside of my monitor screen, I cannot begin to express how grateful I am for the time I’ve been spending at the lab. I remember very clearly the first question he asked me during my interview: “Why should I even listen to you?” Frankly, I had no good answer, and I thought that the meeting didn’t go as well as I’d hoped. Nevertheless, he gave me a chance, and everything began from there.
Initially, I got involved with quality assessment of Multiple Sclerosis and Vasculitis 3D MRI images along with Jason and Amar. Here, I got introduced to the many things Dmitrii can complain about taking brain MRI images. Things such as scanner bias, artifacts, types of imaging modalities and prevalence of disease play a role in how we can leverage these medical images in training predictive models.
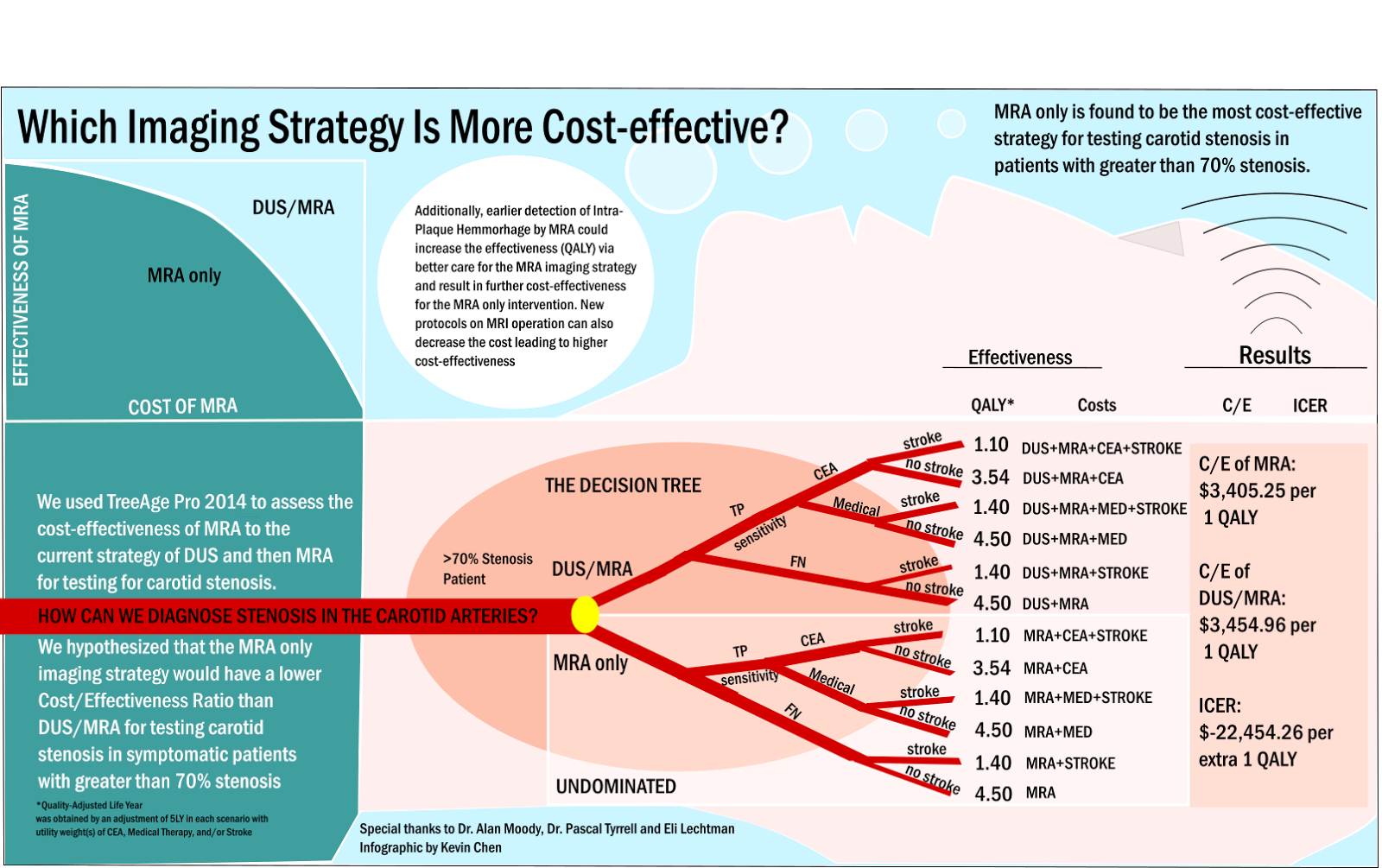
My actual ROP, however, revolved around a niche topic in Mauro and Amar’s project. Their project sought to understand the effect of dataset heterogeneity in training Convolutional Neural Networks (CNN) by cluster analysis of CNN-extracted image features. Upon extraction of image features using a trained CNN, we end up with high-dimensional vectors representing each image. As a preprocessing step, the dimensionality of the features is reduced by transformation via Principal Component Analysis, then selecting a number of principal components (PC) to keep (e.g. 10 PCs). The question must then be asked: How many principal components should we use in their methodology? Though it’s a very simple question, I took way too many detours to answer this question. I looked at the difference between standardization vs. no standardization before PCA, nonlinear dimensionality reduction techniques (e.g. autoencoder) and comparisons of neural network image representation (via SVCCA) among other things. Finally, I proposed an equally simple method for determining the number of PCs to use in this context, which is the minimum number of PCs that gives the most frequent resulting value (from the original methodology).
Regardless of the difficulty of the question I sought to answer, I learned more about practices in research, and I even learned about how research and industry intermingle. I only have Professor Pascal to thank for always explaining things in a way that a dummy such as me would understand. Moreover, Professor Pascal always focused on impact; is what you’re doing meaningful and what are its applications?
I believe that the time I spent with the lab has been worthwhile. It was also here that I discovered that my passion to pursue data science trumps my passion to pursue medical school (big thanks to Jason, Indranil and Amar for breaking my dreams). Currently, I look towards a future, where I can drive impact with data; maybe even in the field of personalized medicine or computational biology. Whoever is reading this, feel free to reach out! Hopefully, I’ll be the next Elon Musk by then…
Transiently signing out,
Stanley Bryan Z. Hua






.jpg)