Hi! My name is Jacqueline, and I’m going into my second year at U of T, pursuing a major in Computer Science and a specialist in Bioinformatics. This past summer, I had the opportunity to do a STA299 project with Professor Tyrrell through the Research Opportunity Program, and I’m excited to share my experiences here!
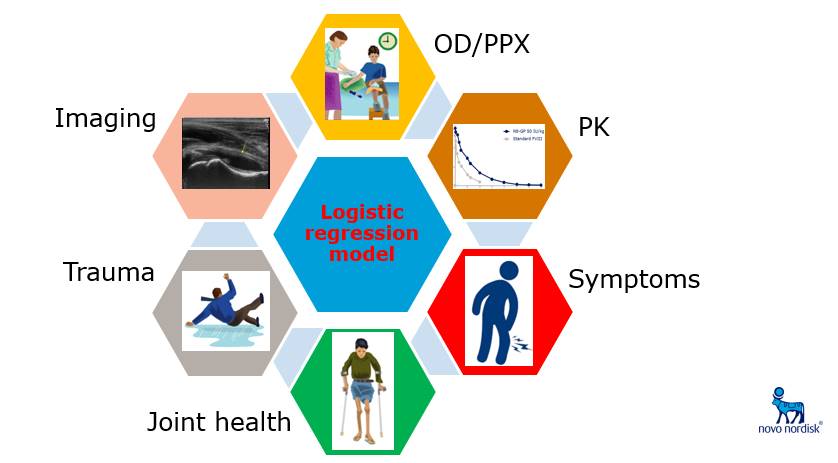
My ROP project for the summer dealt with the simulation of clinical variables relevant to detecting intra-articular hemarthrosis – basically bleeding into the joint – among patients with hemophilia, a disease where patients lack sufficient clotting proteins and are prone to regular, excessive bleeding. Since hemophilia is quite rare, clinical data is often unavailable and so simulation can help us understand what the real data might look like under different sets of plausible assumptions. The ultimate goal was to demonstrate that adding clinical data to Mauro’s existing binary CNN classifier for articular blood detection could boost model performance, as compared to a model trained exclusively on ultrasound images.
Having just completed first year, I went into this ROP with a very limited statistics background and was initially overwhelmed by all the stats jargon being used in lab meetings and in conversations with other lab members. Concepts like “odds ratios,” “ROC,” and “sensitivity analysis” were completely new to me, and I spent many hours just familiarizing myself with these fundamentals.
After a bit of a slow start, I began my project by identifying physical presentation and clinical history variables to use in my simulation. I was fortunate enough to speak with two distinguished hematologists from Novo Nordisk, Drs. Brand-Staufer and Zak, about the features most relevant to diagnosing a joint bleed. Based on this conversation, I selected two of these variables as a starting point and simulated them according to assumed distributions. Next, I simulated the probability of an articular bleed based on a logistic regression model and used that probability to simulate the “true” presence of a bleed based on a Bernoulli distribution.
Then, I took a bit of a fun detour: figuring out how to best match simulated data to real-world bleed probabilities output by Mauro’s model. With some guidance from Professor Tyrrell, I developed a matching algorithm that allowed us to control the strength of the positive correlation between clinical simulated probabilities and classifier probabilities. Perhaps the most difficult part of my project was ensuring that the simulated dataset captured the desired relationships between my explanatory variables and between each explanatory variable and the response variables. Thanks to the advice of Guan and Sylvia, however, I was able to verify these relationships and report on them in a statistically sound manner.
Despite all the obstacles I encountered along the way, despite changing the details of my methodology several times, despite making slow progress and occasionally feeling like I was going in circles, I’m very grateful to have had this opportunity. Not only did I gain a greater understanding of important statistical concepts and greater familiarity with machine learning techniques, but I also got first-hand experience navigating the research process, from beginning to end. Ultimately, my experience in the MiDATA lab was simultaneously challenging and rewarding, and I would like to thank Dr. Tyrrell for all his guidance this summer – whether it was setting up impromptu meetings to discuss unexpected issues in my data, providing feedback on my results, or simply sharing humorous anecdotes in our weekly lab meetings. Regardless of where this next year takes me, I’m confident that I’ll carry the lessons I learned this summer with me.
Jacqueline Seal